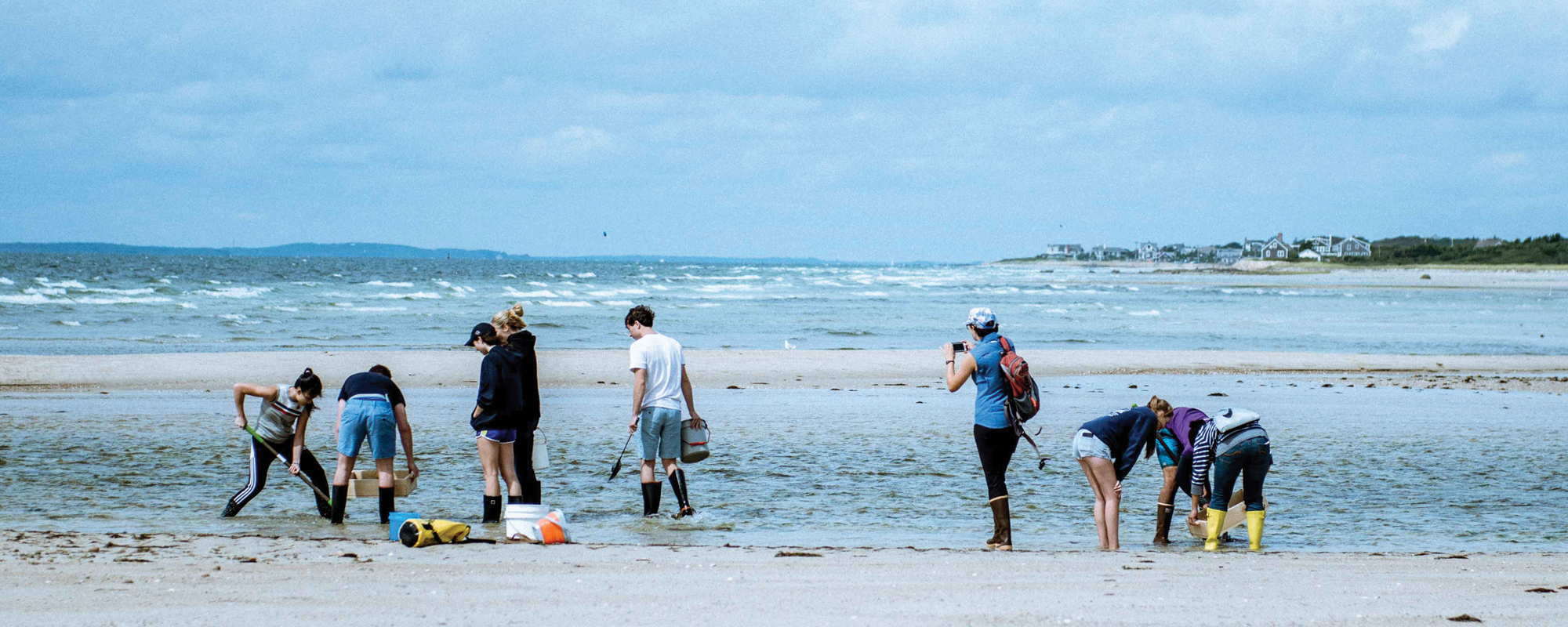
University of Chicago students collect samples for Michael LaBarbera’s course Marine Invertebrates of Woods Hole. (Photography by Megan Costello)
In three weeks, there are just over 500 hours. The students in the Marine Biological Laboratory’s September intensive courses tried to use them all.
In September 2017, the College offered its first three courses at the Marine Biological Laboratory in Woods Hole, Massachusetts. Taught by UChicago and MBL faculty, the intensive three-week courses are designed for students with a strong interest in research. The classes are scheduled to meet five or six days a week, eight hours per day—but most days run much longer.
By the second day of the second week of Jack Gilbert’s course on microbiomes, time is starting to feel a little bent and fuzzy. Gilbert and his group—co-instructor David Mark Welch; teaching assistant Sophia Carryl, SM’17; and a dozen undergraduates—have spent most of the past 24 hours in the lab, working through the night on research projects begun only three days earlier. By the end of next week, those projects will have to be finished.
Meanwhile, last week seems like a distant memory: introductory lectures from Gilbert, a microbial ecologist at UChicago and Argonne, and Mark Welch, an MBL evolutionary biologist, on ecology and microbes and microbiomes, how to investigate them, why they matter. (Some of the students are biology majors, but others come from economics, mathematics, computer science.)
“And then we just gave them carte blanche to come up with research projects,” says a bleary Gilbert, making himself another cup of black tea in the laboratory kitchen.

Students sampled the bacteria in the water and sand and wetlands around MBL; they sampled their own feces and skin. One student, researching whether tidal changes altered the microbiomes on the beach and in the surf, rose before five o’clock one morning to sample the sand at low tide. Another is studying how well bacteria from her fingers survive on coins of different metals—a nickel, a penny, a dime—and whether that microbial residue could be used to identify her. Another is comparing the microbiomes of newly hatched skates to those of adults.
Last night and into this morning, the students extracted the DNA from their many samples and ran polymerase chain reactions to amplify it. It was the first time most of them had attempted either of these procedures. Now, at about 2 p.m. on Tuesday, a few students are trickling back from lunch; the rest are in the lab already, mixing gels and hovering near Carryl, a third-year graduate student whose own research examines avian microbiomes and who was the last one out of the lab the night before. She’s not sure what time that was.
In the room next door, Mark Welch sits down beside a large tabletop device that looks like it might be about to lift off, white and semi-diamond-shaped and pointing toward the ceiling. Accompanied by a student in a blue polo shirt and khaki cargo shorts, he explains how the ultraviolet light inside the machine will activate the dye binding to fragments of DNA, which have been placed on a sheet of milky translucent agarose gel. If everything works—if the DNA sample was extracted properly and the polymerase chain reactions amplified it—then on the computer screen next to the device, “we should see a series of bright bands,” he says. If the results are good, the next step will be sending the DNA off for sequencing to identify the bacterial species.
Mark Welch hesitates for a beat. “OK,” he says. “This is the moment.” He flips the switch to turn on the ultraviolet light. Against the dark background of the computer screen, two rows of white bands appear, some sharp, some ghostly, but all unmistakable. “Ah!” he exclaims. “Hoo-hoo! Oh, that’s beautiful.”
Now it’s closing in on 3 p.m., and everyone trundles upstairs for three student presentations of scientific papers: the resilience of soil microbe diversity, the influence of architectural design on a hospital’s microbiome, the microbial “biogeography” of public restrooms. Being able to tell a compelling story about ideas and findings, Gilbert believes, is an important part of science. For his students, this is practice. It’s also a chance for them to poke around in published papers to look for holes.
“Did they not measure the toilet plume?” one student asks at the end of the restroom study. “You know, like when you flush the toilet and bacteria from inside it go into the air?”
“That’s actually a misnomer,” Gilbert pipes up from the back of the classroom.
“Really?”
“Yeah. Doesn’t actually happen.”
“I’m glad to hear that.” But there is incredulity in her voice.
Gilbert again: “There are so many bacteria on all the surfaces anyway, that we find no more bacteria associated with human stool in the bathroom than we did in the kitchens of homes. When you flush your toilet, you do get aerosolization, but you’re shedding more microbes into your environment than your toilet is generating.” That news comes as less of a comfort.

The paper on architectural design raises other thorny questions, and, guided by Gilbert and Mark Welch, the class uncovers errors of procedure and analysis. “This is a great example of a paper where the authors had a really neat idea, and they went in and biased their entire interpretation,” Gilbert says. “This is an important thing: never color your vision. Take it where the data leads you, not where your impressions and biases lead you.”
Afterward, the group heads back downstairs, where Carryl has been working through the rest of the students’ extracted DNA fragments, imaging everything in the ultraviolet lightbox. Most of the samples came through, she reports; a few did not. Some students will have to try re-extracting the DNA. “This is great,” says Gilbert, who feared that none of the DNA samples might work. Mark Welch concurs: “If this were a real experiment, you might spend multiple days optimizing this whole process”—the extraction, the polymerase chain reactions—“before doing it.”
“And when he says multiple days, he means 30 or 40,” Gilbert adds. He is almost laughing with delight.

They clamber out of the van in borrowed hip waders and rubber boots, chattering with nervous excitement, carrying tools they don’t quite know what to do with yet: shovels and buckets and heavy wood-framed sieves for sifting through the sand that suddenly lay before them on all sides. “All right,” calls Michael LaBarbera, UChicago biology professor emeritus, enunciating over the wind. “This is an exploration—anything
you find out here is fair game.”
It’s a little past 1:30 on a Thursday afternoon, day four of LaBarbera’s course on marine invertebrates, and the tide is falling at Little Sippewissett salt marsh, on the jagged eastern shore of Buzzards Bay, a 10-minute drive from MBL. LaBarbera and his dozen students will spend the next couple of hours here, collecting live creatures to bring back to the lab to study and identify before returning most of them to the ocean a few days later.
Several students head first toward the beach; the rest turn inland, toward the heavy-husked marsh grasses and fingers of brackish water winding back toward the distant woods. LaBarbera goes inland too. He demonstrates how to dig under the sand, scoured clean by the current, to the muckier and darker soil just beneath. That’s where they’d find many more of the worms and mussels and snails they are looking for. Plus the nemerteans and hydrozoans and polychaetes and anomurans on the list LaBarbera handed out the first day of class. “Keep taking samples, even if it seems like there isn’t much,” he says.
“I think you’ll discover you’re getting more organisms than you think.”
He herds the students farther from the beach, deeper into the estuary, where they come upon colonies of fiddler crabs, slipping noiselessly into their burrows as soon as human shadows approach, and larger green crabs, an invasive species from Europe, LaBarbera explains, which came to America on the hulls of early 1800s ships.
One student reports gleefully that a handful of tiny clear shrimp jumped into her bucket when she stooped to fill it with water. Another reaches into the creek and lifts out an empty, translucent, perfectly intact shell, left behind by a molting crab. Another finds a baby horseshoe crab, barely a month old and smaller than a thumbnail. LaBarbera has never seen one so young and tiny. “Newly metamorphosed,” he says. He will come back in a few days to return it to this exact spot to give the crab its best chance of survival. “If it gets through this next year, this guy can live another 20 years. Some things just pull your heartstrings.”

The students keep wading upstream. They find a rock covered in barnacles with an oyster attached, very much alive, with its shell tightly clenched.
Someone finds a softshell blue crab. “Oh gosh!” The whole group comes splashing over. LaBarbera tells them the crabs often mate when females are molting—“partly because sperm transfer is more efficient then”—and afterward, the male will cradle the female in his legs, carrying her for the next 48 hours, while her new exoskeleton hardens. “Then they go their separate ways. You can view this as the male protecting his reproductive investment,” LaBarbera says, grinning slightly, “or you can view it as the only moment of romance in a blue crab’s life.”
Another find. A student wades over to LaBarbera, holding out what looks like a tiny ice cream cone, two inches long and thinner than a straw. “Oh!” he says. “Anybody know what this is?”
“It looks like an egg case?” the student guesses.
“Nope,” LaBarbera says. “It’s a tube. The animal inside it—see in there?—that’s a worm. And it constructs this out of sand.” Each tube is one grain thick, built piece by piece like a brick wall. “The most amazing structure you’ve ever seen,” LaBarbera says. “We’ll take it back to the lab and you can look at it under the scope.” The worm, meanwhile (a Pectenaria, though LaBarbera doesn’t give the name away—he wants the students to find it), is “astonishing,” he confides, after the others wade ahead. “It has these little chitin setae, these little hair-like structures that stick out the front, and they’re this brilliant golden color.” They look, he says, “ like a golden beard.”
By now it’s getting close to 3:45, when the van will return to haul the group and their buckets full of specimens back to MBL, where the students will stay in the lab long past the official 5:30 p.m. quitting time, until 9 or 10, combing through reference books and lists of species, looking for what they had found, putting names to the nameless. They'll discover organisms in their buckets that they hadn’t realized they’d collected: sea snails as small as specks of dirt, a worm with a missing back end, an impossibly small anemone.
LaBarbera watches them wind their way toward the back of the estuary, bending to shovel and sift the sand, then wading on a little further. He doesn’t want to summon them back just yet. “The longer they’re out here,” he says, “the more they’ll see.” He wants them to see it all.

On the day before their last day at MBL, the students in neurobiologist Eric Schwartz’s course on proteins are huddled over microscopes and amplifiers and culture dishes, working to complete the laboratory projects begun barely 48 hours earlier—each one wound around a slightly different molecular mystery. Tomorrow afternoon, they’ll present their findings at an open house finale for the three September courses. The morning after that, they’ll all be en route back to Chicago. In one of the closet-sized chambers at the back of the lab, a pair of students is sending different voltage pulses into a frog oocyte, an immature and unfertilized egg cell, to see whether the proteins they injected into its membrane would fluoresce. Minute and bead-like and nut-brown, the oocyte, attached to two glass electrodes and suspended in a beam of light, sits atop a microscope that the students built themselves. There are lenses and mirrors and a photodiode to measure how much light comes off the cell.
Three days ago, they hadn’t known how to assemble these parts into a working instrument. “It was kind of a steep learning curve,” says one student, a young man in shorts and glasses and a gray MBL hoodie. “More like learning skydiving than anything.” But now here he is, talking about the precise placement of the mirrors and the LED that will shine a light of a very specific wavelength, 470 nm—“very pure, very blue”—onto the oocyte. He explains that if the cell lights up then they’ll know that the protein they’ve injected was a genetically encoded voltage indicator: “The more voltage you put in, the more it fluoresces.”
The course, called Observing Proteins in Action: How to Design and Build Your Own Instruments, is really two courses, or maybe three. The students learn how to put together microscopes and amplifiers, after two weeks of lessons on optics and electronics. But they also learn the basics of proteins: how they function and influence the behavior of cells and organisms, how to isolate certain proteins and incorporate them into a cell membrane, how to monitor their activity, millisecond by millisecond.
Schwartz, a professor in the Department of Pharmacological and Physiological Sciences, coteaches the course with three UChicago colleagues from the Department of Biochemistry and Molecular Biology: Ana Correa, Francisco Bezanilla, and Eduardo Perozo. One of the guiding concepts for the syllabus, Schwartz says, is a simple not-so-simple question: “How do we know what we know?” The answer, or one answer, is instruments—like microscopes. But that’s not simple either. How scientists modify instruments determines what things they can discover. “It changes what we know,” said Schwartz, who first came to MBL as a medical student 50 years ago, when he spent two summers as a research assistant. “The idea here was to have the students put everything together on their own, so they can understand not just what they’re measuring, but how.”
In another little room off the lab, two young women, with their own hand-built microscope, are testing two potassium-blockers—they haven’t been told exactly what they are—to find out how the drugs work, by what mechanism they interfere with the oocyte’s potassium ion channel. The experiment involves sending voltage into the cell and reading the current that comes out. The students are watching to see how the bright green line on a graph on their laptop screen moves. “That’ll tell us what is being expressed in our frog egg with the RNA we inserted,” one student says. “That’s how we’ll know.”
In the building next door, two other students are sitting in the cool dark hum of the MBL’s electron microscope, its long white tower of lenses rising from the desktop like a smokestack, or a periscope. The previous day, the students isolated and purified ribosomes—the cellular factories where proteins are made—from E. coli, then stained them. Now, under the electron microscope, with the magnification dialed up to 100,000, then 150,000, then 200,000, the ribosomes spring onto the computer screen in crisp black and white, perfectly formed Tetris-like shapes frozen in a dark sea. “That’s beautiful,” says teaching assistant Michael Clark, an MD/PhD student in the Pritzker School of Medicine. “Oh my goodness. Good job, guys.”
Meanwhile, in the lab, the course’s only nonscience student—“I’m a humanities major,” she says, with a pleading laugh—is toiling away on a circuit to measure the dropping voltage across a very small area of a Venus flytrap after the plant’s trigger hairs are touched. “Because they’ve got these tiny hairs, right?” she explains. “And they’re super sensitive. And if you touch them, it triggers the opening of some ion channels, which will basically—see, one side of the cell membrane has more calcium than the other, and if you open the ion channels, the calcium floods in, and the water pressure in the plant goes way up, and the plant slams shut.”
Amid that furious rush, the voltage in the flytrap’s membrane shifts. “What I’m trying to measure is that change in voltage.” She adjusts her glasses and picks up a blue marker to draw an explanatory diagram in a notebook full of crossed-out diagrams (“This is like six drawings of the same circuit, all drawn in slightly different wrong ways.”).
She’s been at this work for hours, cheerfully, patiently, arranging switches and resistors and capacitors and amplifiers and function generators on a breadboard—a unit for building a simple electronic circuit without requiring any soldering. When everything else is ready, an electrode connected to a pipette holder will carefully touch the trigger hairs and conduct the charge from the excited plant to the circuit. “It’s going to be a teeny tiny signal,” she says. “If I get any measurement at all, I’ll be pleased.”
Now dinnertime is approaching. It's going to be a long night for all of them. But after three weeks, they are used to that; they know how to pace themselves. The humanities major glances up at the big round clock on the back wall. It would carry some of them into the morning.
