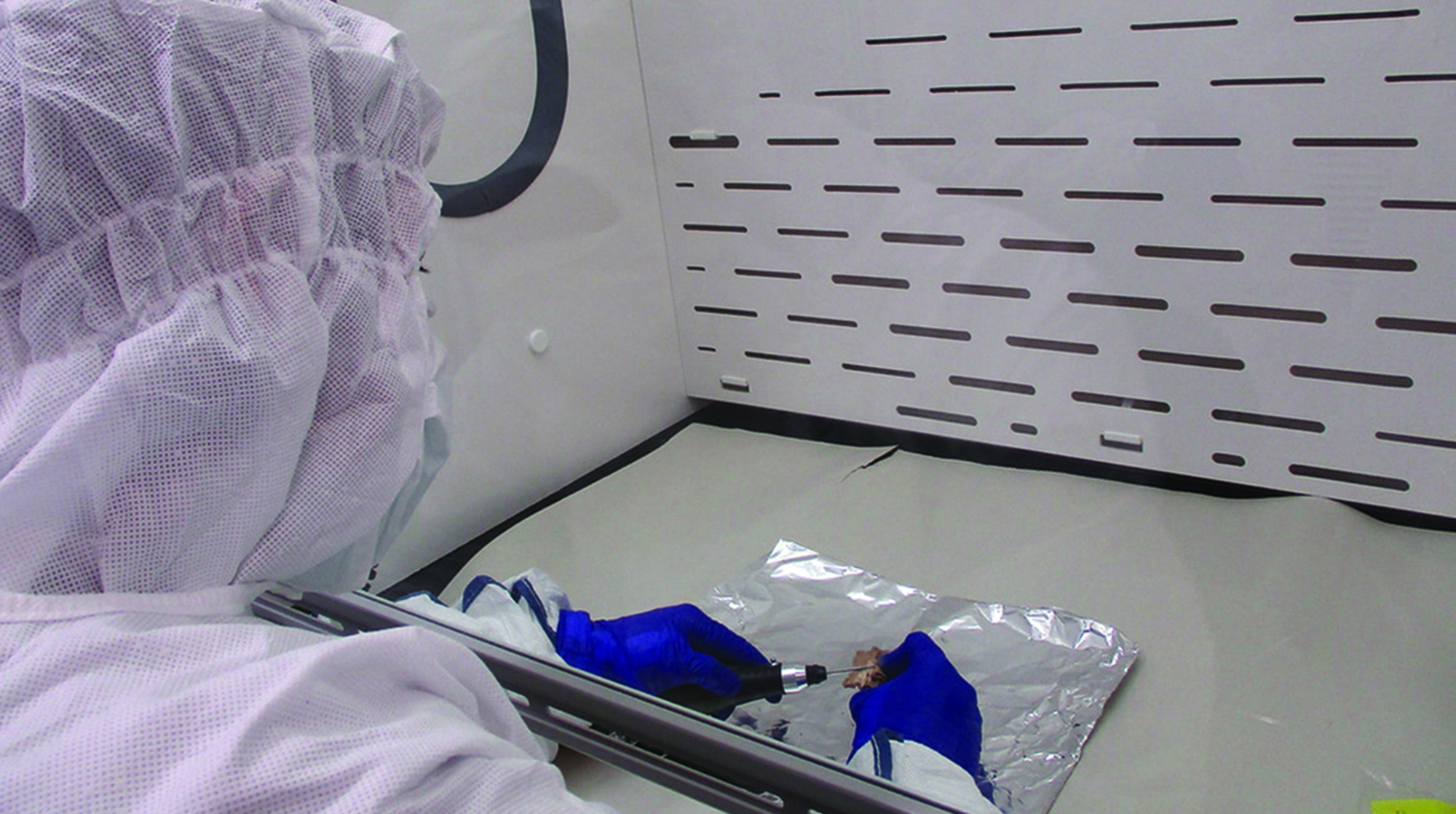
PhD student Esha Bandyopadhyay, under a fume hood, uses a sterile drill on a petrous bone from an ancient human. The DNA in this inner ear bone is better preserved than that in other bones. The resulting powder is used for DNA extraction and sequencing. An ancient human petrous bone. (Photography by Constanza de la Fuente)
Paleogenomicist Maanasa Raghavan maps ancient DNA to chart human history.
It was 2006. Maanasa Raghavan had just graduated from the University of Toronto with a bachelor’s degree in molecular biology when she and her friends headed off to Europe. They were touring not the grand cathedrals nor the ancient forums, but places further into the mists: archaeological sites dating back tens or hundreds of thousands of years. Though she had an abiding love of history and archaeology, Raghavan was unsure how to make a career of it. In fact, come autumn she had plans to begin a PhD in cancer research back in Canada.
Plans changed. While in Europe, Raghavan and her friends volunteered for a time at Cova Gran, a Paleolithic rock shelter in northeast Spain. There they encountered a group of researchers who told them about a recent paper analyzing DNA extracted from a Neanderthal fossil. At first Raghavan thought they were kidding: “I had no idea you could actually get DNA from ancient people,” she says. When she realized they were serious, it “suddenly opened the door on a path that would marry my knowledge and training in genetics with my interest in history and archaeology.”
Raghavan then earned her master’s degree in archaeological sciences at the University of Oxford and her doctorate in paleogenomics at the University of Copenhagen; most labs at the time were based in Europe. Now a Neubauer Family Assistant Professor of Human Genetics at UChicago, she is using the blueprint of genetics to piece together how people have moved and mingled through history. Her primary focus is on the past 10,000 years, connecting the advent of agriculture and, later, the settlement of cities to the flow of people and their genes around the globe.
The field of paleogenomics, as the study of ancient DNA is known today, was born in 1984, when researchers from the University of California, Berkeley, and the San Diego Zoo traveled to a museum in Mainz, Germany, to collect traces of dried muscle from the preserved skin of a quagga—a zebralike animal that had gone extinct a century earlier. By mapping part of the genome, the team demonstrated for the first time that DNA can be collected and analyzed long after an animal’s death. They used what they found to trace the quagga’s evolutionary link to zebras.
Of the two kinds of DNA, nuclear and mitochondrial, the latter was the kind available to these earliest researchers. Being passed down only through the maternal line, the mitochondrial genome yields less information than the nuclear; but, because each cell contains one nucleus and many mitochondria, more of the latter survived over time.
The field grew in fits and starts, then bloomed in the 2000s as sequencing technologies improved and standards for managing degradation and contamination in ancient samples were codified.
“Prior to these methods, we would take DNA from contemporary populations and then try to extrapolate back in time using population genetics and statistical methods to get a sense of past genetic variation,” Raghavan says. “We don’t have to use those assumptions anymore. We have the empirics to ground our inferences.”
What has come from this work—Raghavan’s and others’—is the refinement and reevaluation of some historical narratives. It had been believed, for instance, that farmers who migrated from the Middle East several millennia ago replaced hunter-gatherer populations in some parts of Europe and interbred with them in others. Starting in 2010, however, studies using ancient DNA were able to investigate the nuclear genome and to produce a more complex picture in which the hunter-gatherer societies were not replaced but rather folded into at least two waves of settlers, first the farmers and then groups of pastoralists from the Caspian steppe. This work is done by sampling DNA from each of these populations and then comparing the genetic fingerprints to determine whether, and to what degree, they lived together and mixed their gene pools to give rise to present-day Europeans.
The uniquely human ability to metabolize milk after weaning is another puzzle that ancient DNA has helped crack. Researchers have generally assumed that this ability, rooted in human production of the lactase enzyme, emerged around the same time as agriculture. The logic, says Raghavan, is clear: we domesticated cattle, and then began milking them. But more recent inquiry using ancient DNA suggests instead that the Caspian pastoralists who migrated into Europe about 5,000 years ago—the Yamnaya culture—possessed the ability to digest milk. Given milk’s dense nutritional value, this particular genetic trait dispersed rapidly.
As paleogenomics has expanded and well-funded labs have become more common, ethical questions have grown in lockstep. How, for example, can the physical collection of ancient individuals be done sustainably and respectfully? As Raghavan noted, many of them are ancestors of present-day Indigenous peoples who have been subject to centuries of exploitation.
“The push for high-profile publications, coupled with the destructive sampling needs of paleogenomics, has driven extractive and unethical practices,” she says. “Ambition can prevent people from thinking about the damage being done.” Though Raghavan’s lab relies as much as any other on these methods, she and several colleagues are publicly tackling the question of how to most respectfully and dutifully handle such a limited and precious resource.
They are also thinking through broader ethical concerns rooted in histories of colonial extraction. How can Indigenous populations and local researchers be involved in framing and conducting the research? Can these projects provide more explicit benefit to the populations in which they’re rooted, rather than generating a wholly extractive form of knowledge, stashed behind the paywalls of academic journals?
As the field of paleogenomics emerges from adolescence and these matters of ethics are resolved, Raghavan is focused on projects in India, Sri Lanka, Pakistan, Turkey, and South America. She is tracking down ancient traces of adenine and thymine, cytosine and guanine, and from these drafting a sentence or two in the story of humankind.
